our projects
Our group is focused on improving outcomes for patients diagnosed with early-stage colon and rectal cancer.
There are >1.4 million cases of colorectal cancer (CRC) diagnosed annually worldwide. Due to national screening programmes, the predominant stage of CRC diagnosis has recently shifted from late- to early-stage. Despite having better prognosis when compared to metastatic disease, 1 in 3 patients with surgically resectable stage II/III CC will recur with liver metastasis, where survival rates remain <10%. Multiple molecular and histological profiling studies, including our own, have identified patients most at risk of this metastatic relapse have tumours characterised by the presence of early disseminating cells in the form of tumour buds, elevated signalling associated with activation of the TGF-β pathway and overall histological appearance that is distinguishable due to their high stromal (particularly fibroblast) content.
Such “stroma-rich” tumours with high levels of tumour buds have a metastatic recurrence rate of approximately 50%; resulting in this group being prioritised for treatment intensification. However, despite advances in treatment options, multiple trials and meta-analyses have demonstrated that patients with stroma-rich tumours derive minimal benefit from standard-of-care chemotherapies, reinforcing the importance for the need for new therapeutics for this poorly served clinical group.
Our group is working on a number of projects that aim to identify the biology driving aggressive features in early stage CRC, and using this new understanding to develop and test new treatment options for patients. To do this we utilise multi-omics and histological data from human colon tumours (n=2400 patients samples) and GEMMs (n=110 from n=22 genotypes) available to the Dunne group via consortiums like S:CORT and ACRCelerate.
Research projects within the group:
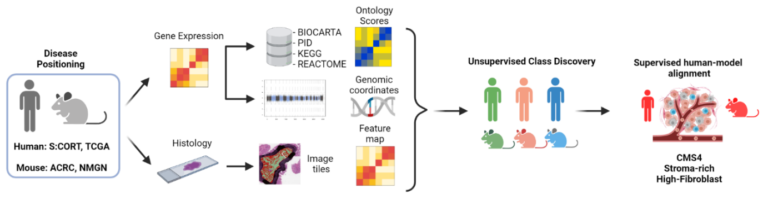
Generate a dual-species “deep phenotyping” systems (Belfast, Beatson, Oxford):
Triangulating information from transcriptional, histological, and mutational alignment across mouse/human tumours will provide human subtype-matched classification of mouse tumours enabling selection of the most appropriate models for pre-clinical testing. Dual-species deep phenotyping characterisation is being performed using data already established within the S:CORT and CRUK-funded ACRCelerate programmes. Development of dual-species disease-positioning approaches, where comparative deep phenotyping of GEMMs is performed in parallel with human tumours, enable accurate description of the cancer-host interactions, providing a more translatable platform to test therapeutic options and predict clinical response.
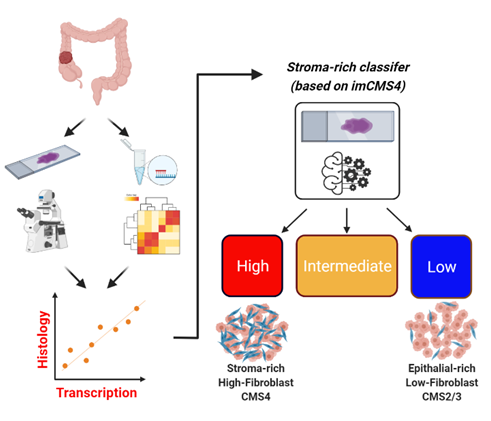
Develop and validate rapid turnaround digital pathology-based classifiers (Belfast, Oxford, Zurich):
A major limitation in translating RNA-based signatures is the weeks needed to extract and profile the RNA; a timeline incompatible with clinical decision-making. Using digital pathology capabilities and expertise across our collaborative network, alongside tumour tissue routinely collected, we have been developing semi-automated digital-pathology companion diagnostic classifiers.
As part of the MRC-funded S:CORT programme, our team has access to digital images from numerous clinical trials; using this existing image data, we have recently developed an algorithm to identify “stroma-rich” tumours, which we are refining to ensure if can be used to stratify patients in a rapid and robust way. With increased use of digital pathology for diagnostic reporting, the classifiers we are developing are being designed in a way to align with diagnostic pipelines, ensuring patient selection maintains high standards and reporting uniformity.
A major limitation in translating RNA-based signatures is the weeks needed to extract and profile the RNA; a timeline incompatible with clinical decision-making. Using digital pathology capabilities and expertise across our collaborative network, alongside tumour tissue routinely collected, we have been developing semi-automated digital-pathology companion diagnostic classifiers.
As part of the MRC-funded S:CORT programme, our team has access to digital images from numerous clinical trials; using this existing image data, we have recently developed an algorithm to identify “stroma-rich” tumours, which we are refining to ensure if can be used to stratify patients in a rapid and robust way. With increased use of digital pathology for diagnostic reporting, the classifiers we are developing are being designed in a way to align with diagnostic pipelines, ensuring patient selection maintains high standards and reporting uniformity.
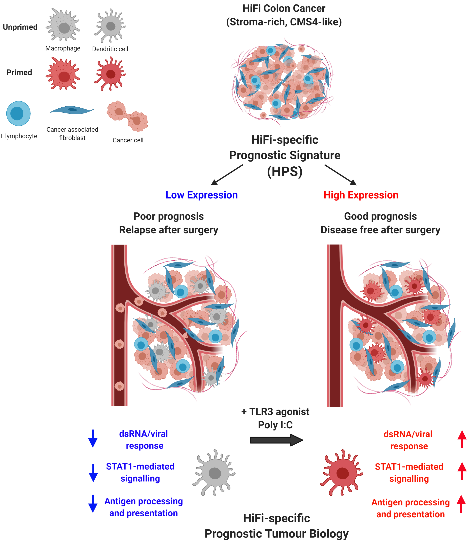
New treatment options for stroma-rich CRC (Belfast, Glasgow, Oxford, NCRI):
To address the lack of efficacious therapeutic options for patients with stroma-rich CRC, our group have histologically stratified a series of independent human tumour cohorts according to stromal content, enabling identification of biology underpinning relapse and potential therapeutic vulnerabilities specifically within stroma-rich tumours that could be exploited clinically. Utilising in silico, in vitro and in vivo models, this work has demonstrated the subtype-specific prognostic value of antigen processing and presentation within discrete immune lineages in stroma-rich CC; signalling that is stimulated following treatment with the dsRNA and TLR3 agonist poly(I:C), resulting in significantly reduced liver metastases in an in vivo model. Our group is working within the national NRCI clinical networks to translate these findings into a clinical trial.
Risk stratification in very early-stage colorectal cancer (Belfast, Utrecht, Oxford, Leeds, Sanger, Birmingham):
Bowel cancer screening has led to an increase in the number of CRCs being diagnosed at an early stage. Although typically an early diagnosis correlates with good response rates to treatment, some patients have ‘born-to-be-bad’ tumours which progress to metastatic disease despite early diagnosis. Currently we are unable to distinguish between tumours with a good prognosis and those that are ‘born-to-be-bad’ at the time of diagnosis. Our CRUK-funded project aims to investigate ways to identify tumours that are at high risk of early dissemination through molecular profiling, thus guiding clinical decision making.
Research tools developed by the group:
Interpretation of molecular signatures (Belfast, Glasgow, Oxford, Zurich):
Research within our group has highlighted that heterogeneity between tumour samples, such as differences in the proportions of epithelial, immune and stromal cells, has the potential to confound our understanding and interpretation of gene signatures that are applied to these samples. For example, signatures developed to characterise mesenchymal traits within epithelial cell lines, when applied to bulk tumour data, are likely to be enriched within non-tumour epithelial cell types, and therefore become highly-accurate tumour stromal percentage estimates rather than measures of subtle epithelial transitions.
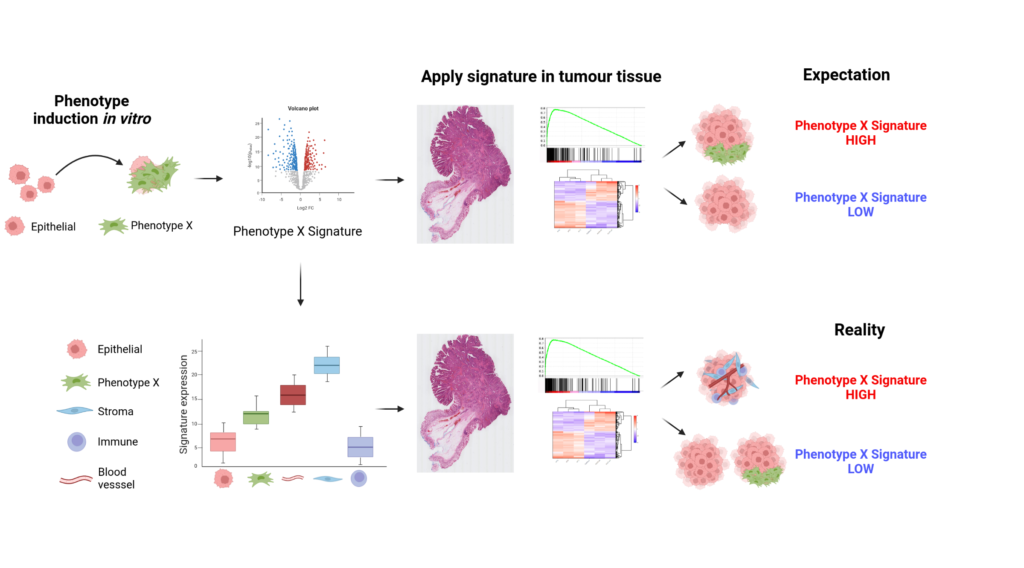
As a result of this we suggest that caution should be applied when interpreting the biological significance of gene signatures being tested. To allow researchers to assess their own gene signatures and gain a more informed interpretation of the true biology and cell-type expression underlying the signatures we have developed the ConfoundR resource, which indicates if the signature being tested could be susceptible to stromal confounding issues. This will then allow researchers to adjust the interpretation of the data, if required, improving patient stratification based on more accurate interpretation of the underlying biology.
The ConfoundR resource can be accessed here: www.confoundr.qub.ac.uk
Easy to use on-line tool for human-mouse computational analysis
Generation of transcriptional data has dramatically increased in the past decade, driving the development of analytical algorithms that enable interrogation of the biology underpinning the profiled samples. However, these resources require users to have expertise in data wrangling and analytics, reducing opportunities for biological discovery by ‘wet-lab’ users with a limited programming skillset. Although commercial solutions exist, costs for software access can be prohibitive for academic research groups.
To address these challenges, we have developed an open source and user-friendly data analysis platform for on-the-fly bioinformatic interrogation of transcriptional data derived from human or mouse tissue, called Molecular Subtyping Resource (MouSR; https://mousr.qub.ac.uk/).
This internet-accessible analytical tool enables users to easily interrogate their data using an intuitive ‘point-and-click’ interface, which includes a suite of molecular characterisation options including quality control, differential gene expression, gene set enrichment and microenvironmental cell population analyses from RNA sequencing. The MouSR online tool provides a unique freely available option for users to perform rapid transcriptomic analyses and comprehensive interrogation of the signalling underpinning transcriptional datasets, which alleviates a major bottleneck for biological discovery.

